Reorientation lifetime of vectors#
from fishmol import trj, utils, funcs, style
from cage_data import cage1_info
Read trajectory file#
%%time
cell = cage1_info.cell
traj = trj.Trajectory(timestep = 5, data = "/nobackup/rhtp48/data_ana/cage1-500K.xyz", index = ":", cell = cell)
CPU times: user 2min 17s, sys: 7.39 s, total: 2min 24s
Wall time: 2min 27s
Vector reorientation dynamics (VRD) of water O-H bonds#
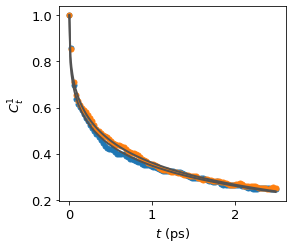
Sepearately calculate the O-H bonds#
water_vrd = funcs.VRD(traj = traj, spec = [14, [15, 16]], num = 500, sampling = 5, skip = 2)
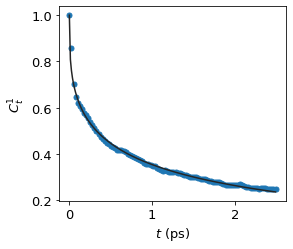
results = water_vrd.calculate(l = 1, mean = False, fit = True, plot = True)
Progress: [■■■■■■■■■■■■■■■■■■■■] 100.0%
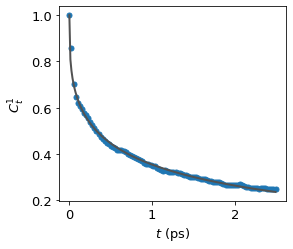
Average the O-H bonds of the same water molecule#
water_vrd = funcs.VRD(traj = traj, spec = [14, [15, 16]], num = 500, sampling = 5, skip = 2)
results = water_vrd.calculate(l = 1, mean = True, fit = True, plot = True)
Progress: [■■■■■■■■■■■■■■■■■■■■] 100.0%
Plot the results by yourself#
fig, ax = plt.subplots()
ax.scatter(results.t, results.C_t)
ax.plot(results.t_fit, results.C_t_fit, color = "#252525")
ax.set_xlabel(r"$t$ (ps)")
ax.set_ylabel(r"$C^1_t$")
plt.show()
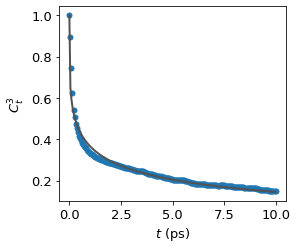
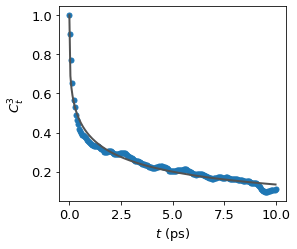
VRD of trifluoroacetate ion C-C bond#
Calculate the C-C bond reorientation dynamics in one TFA anion#
TFAs = cage1_info.TFAs
spec = [*TFAs[0].values()]
spec
[0, 1, 2, 3, 4, 5, 6]
print([x for x in spec if traj.frames[0][x].symbs == "O"])
[3, 4]
vrd = new_VRD(traj = traj, spec = [3, 4], num = 2000, sampling = 10, skip = 5)
results = vrd.calculate(l = 3, mean = False, fit = True, plot = True)
Progress: [■■■■■■■■■■■■■■■■■■■■] 100.0%
all_oxygen = [x for spec in TFAs for x in [*spec.values()] if traj.frames[0][x].symbs == "O"]
specs = [[x for x in all_oxygen[::2]], [x for x in all_oxygen[1::2]]]
specs
[[3, 10, 132, 139, 261, 268, 390, 397], [4, 11, 133, 140, 262, 269, 391, 398]]
Calculate the average VRD of all TFA C-C bonds#
vrd = funcs.VRD(traj = traj, spec = specs, num = 2000, sampling = 10, skip = 5)
results = vrd.calculate(l = 3, mean = True, fit = True, plot = True)
Progress: [■■■■■■■■■■■■■■■■■■■■] 100.0%