MSD and diffusion coefficient#
Import packages#
from fishmol import trj, msd
from cage_data import cage1_info
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from fishmol import style
Import information of the simulated system#
# The simulation box
cell = cage1_info.cell
# Waters is a dict contains the indices of atoms of water molcules
waters = cage1_info.waters
water_indices = [[*water.values()] for water in waters]
print(water_indices)
[[14, 15, 16], [17, 18, 19], [143, 144, 145], [146, 147, 148], [272, 273, 274], [275, 276, 277], [401, 402, 403], [404, 405, 406]]
Read trajectory data file#
%%time
# The trajectory is a large file (517 MB)
traj = trj.Trajectory(timestep = 5, data = "/nobackup/rhtp48/data_ana/fishmol_examples/cage_data/cage1-500K.xyz", index = ":", cell = cell)
CPU times: user 45.4 s, sys: 1.47 s, total: 46.9 s
Wall time: 47.1 s
You can retrieve and confirm the simulation cell info
traj.cell
[[21.2944, 0.0, 0.0],
[-4.6030371123, 20.7909480472, 0.0],
[-0.9719093466, -1.2106211379, 15.1054299403]]
A test of calc_com() function is working
traj.frames[0][water_indices[0]].calc_com()
array([10.58631557, 19.90213351, 10.38000923])
Calculate the centre of masses (CoMs) for water molecules#
from fishmol.utils import update_progress # import the progress bar function
water_coms = np.zeros((traj.nframes, 3*len(water_indices)))
for i, frame in enumerate(traj.frames):
# Calculate com
coms = [frame[water_idx].calc_com() for water_idx in water_indices]
water_coms[i] = np.asarray(coms).flatten()
update_progress(i/traj.nframes)
# Add data to dateframe
columns=[]
for i in range(1, len(water_indices) + 1):
columns += [f"water{i}_x", f"water{i}_y", f"water{i}_z"]
water_com_df = pd.DataFrame(columns=columns, data = water_coms)
# Write data to excel file
water_com_df.to_excel("test/cage1-500K-water-com.xlsx")
update_progress(1)
Progress: [■■■■■■■■■■■■■■■■■■■■] 100.0%
water_com_df.head()
| water1_x | water1_y | water1_z | water2_x | water2_y | water2_z | water3_x | water3_y | water3_z | water4_x | ... | water5_z | water6_x | water6_y | water6_z | water7_x | water7_y | water7_z | water8_x | water8_y | water8_z | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10.586316 | 19.902134 | 10.380009 | 0.624155 | 10.923925 | 8.298079 | -1.967392 | 4.261146 | 12.241481 | 4.693672 | ... | 4.725172 | 15.090375 | 8.649502 | 6.883228 | 17.759366 | 15.321196 | 2.865725 | 11.019290 | 3.578953 | 0.703805 |
| 1 | 10.597616 | 19.945151 | 10.393126 | 0.617881 | 10.942585 | 8.312331 | -1.961926 | 4.253910 | 12.205821 | 4.689873 | ... | 4.745156 | 15.076014 | 8.640429 | 6.849210 | 17.749958 | 15.278667 | 2.889409 | 11.049146 | 3.587807 | 0.663123 |
| 2 | 10.610208 | 20.003821 | 10.408405 | 0.610859 | 10.967922 | 8.330438 | -1.953047 | 4.243309 | 12.156421 | 4.687198 | ... | 4.767198 | 15.058083 | 8.628816 | 6.804086 | 17.738720 | 15.225162 | 2.920655 | 11.086905 | 3.600664 | 0.608757 |
| 3 | 10.622718 | 20.062112 | 10.416761 | 0.604688 | 10.993416 | 8.349495 | -1.945208 | 4.229969 | 12.106099 | 4.687512 | ... | 4.786539 | 15.040753 | 8.617851 | 6.760128 | 17.728982 | 15.181476 | 2.948253 | 11.122864 | 3.614650 | 0.558271 |
| 4 | 10.636664 | 20.122749 | 10.416813 | 0.597829 | 11.020753 | 8.373153 | -1.941190 | 4.211549 | 12.054352 | 4.691621 | ... | 4.808036 | 15.020218 | 8.605213 | 6.714651 | 17.717884 | 15.151479 | 2.970079 | 11.155729 | 3.629731 | 0.511643 |
5 rows × 24 columns
del water_coms
Calculate the mean squared displacement of water molecules#
Create the time data#
start_idx = 0
t0 = start_idx * 5
t_end = t0 + 5*(len(water_com_df)-1) # 5 fs is the interval of traj
t = np.linspace(t0, t_end, num = len(water_com_df))
msd_d_df = pd.DataFrame()
msd_d_df["t"] = t[1:]
Calculate the MSD and D#
Define a function to calculate the mean squre displacements (MSDs).
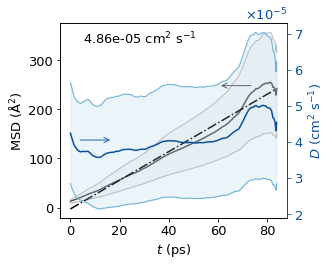
The mean squared displacement (MSD) measures how much particles move over time. There are a number of definitions for the mean squared displacement. The MSDs here are calculated from the most common form, which is defined as: $\( MSD(m) = \frac{1}{N_{particles}} \sum_{i=1}^{N_{particles}} \frac{1}{N-m} \sum_{k=0}^{N-m-1} (\vec{r}_i(k+m) - \vec{r}_i(k))^2 \)\( where \)r_i(k)\( is the position of particle \)i\( in frame \)k\(. The mean squared displacement is averaged displacements over all windows of length \)m$. The algorithm used is described in calandrini2011nmoldyn which can be realised by the code in this StackOverflow thread.
for i in range(len(water_indices)):
temp = np.array(water_com_df.iloc[:, 3*i:3*i+3])
msds = msd.msd_fft(temp)
msd_d_df[f"water{i}_MSD"] = msds[1:]
msd_d_df[f"water{i}_D"] = msds[1:] * 1E-16 / (6*t[1:]*1E-15)
msd_df = msd_d_df.iloc[:, 1:16:2]
d_df = msd_d_df.iloc[:, 2:17:2]
Calculate average values of MSD and D#
msd_d_df["Mean_MSD"] = msd_df.mean(axis=1)
msd_d_df["MSD_error"] = msd_df.std(axis=1) / 8**0.5
msd_d_df["Mean_D"] = d_df.mean(axis=1)
msd_d_df["D_error"] = d_df.std(axis=1) / 8**0.5
# Save the data
msd_d_df.to_excel("test/cage1-H2O-MSD-D-500K.xlsx")
Plot the data#
# skip the first 5 ps, that the system is still equilibrating
msd_d_df = msd_d_df.iloc[1000:,:]
msd_d_df["t"] = (msd_d_df["t"] -5000)
msd_d_df.head()
| t | water0_MSD | water0_D | water1_MSD | water1_D | water2_MSD | water2_D | water3_MSD | water3_D | water4_MSD | ... | water5_MSD | water5_D | water6_MSD | water6_D | water7_MSD | water7_D | Mean_MSD | MSD_error | Mean_D | D_error | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1000 | 5.0 | 4.093919 | 0.000014 | 19.722342 | 0.000066 | 2.003004 | 0.000007 | 33.566337 | 0.000112 | 1.458750 | ... | 19.347635 | 0.000064 | 2.614106 | 0.000009 | 19.172680 | 0.000064 | 12.747347 | 4.198995 | 0.000042 | 0.000014 |
| 1001 | 10.0 | 4.095739 | 0.000014 | 19.746275 | 0.000066 | 2.003836 | 0.000007 | 33.597794 | 0.000112 | 1.458538 | ... | 19.358398 | 0.000064 | 2.613060 | 0.000009 | 19.178147 | 0.000064 | 12.756473 | 4.202892 | 0.000042 | 0.000014 |
| 1002 | 15.0 | 4.097586 | 0.000014 | 19.770177 | 0.000066 | 2.004774 | 0.000007 | 33.629258 | 0.000112 | 1.458261 | ... | 19.369117 | 0.000064 | 2.612067 | 0.000009 | 19.183779 | 0.000064 | 12.765627 | 4.206788 | 0.000042 | 0.000014 |
| 1003 | 20.0 | 4.099463 | 0.000014 | 19.794038 | 0.000066 | 2.005816 | 0.000007 | 33.660730 | 0.000112 | 1.457916 | ... | 19.379795 | 0.000064 | 2.611117 | 0.000009 | 19.189575 | 0.000064 | 12.774806 | 4.210683 | 0.000042 | 0.000014 |
| 1004 | 25.0 | 4.101374 | 0.000014 | 19.817852 | 0.000066 | 2.006957 | 0.000007 | 33.692212 | 0.000112 | 1.457499 | ... | 19.390432 | 0.000064 | 2.610204 | 0.000009 | 19.195537 | 0.000064 | 12.784008 | 4.214578 | 0.000042 | 0.000014 |
5 rows × 21 columns
fig = plt.figure(figsize=(4.6,3.6))
ax = fig.add_axes([0.16, 0.16, 0.685, 0.75])
color = "#08519c"
ax.plot(msd_d_df["t"]/1000, msd_d_df["Mean_MSD"], color = "#525252")
ax.plot(msd_d_df["t"]/1000, msd_d_df["Mean_MSD"] - msd_d_df["MSD_error"], linewidth = 1, color = "#bdbdbd")
ax.plot(msd_d_df["t"]/1000, msd_d_df["Mean_MSD"] + msd_d_df["MSD_error"], linewidth = 1, color = "#bdbdbd")
ax.fill_between(msd_d_df["t"]/1000, msd_d_df["Mean_MSD"] + msd_d_df["MSD_error"], msd_d_df["Mean_MSD"] - msd_d_df["MSD_error"], color= "#d9d9d9", alpha = 0.2)
linear_model = np.polyfit(msd_d_df["t"]/1000, msd_d_df["Mean_MSD"],1)
linear_model_fn = np.poly1d(linear_model)
x_s = np.arange(msd_d_df["t"].min()/1000, msd_d_df["t"].max()/1000)
ax.plot(x_s,linear_model_fn(x_s), color="k", ls ="-.")
ax1 = ax.twinx()
ax1.plot(msd_d_df["t"]/1000, msd_d_df["Mean_D"], color = color)
ax1.plot(msd_d_df["t"]/1000, msd_d_df["Mean_D"] - msd_d_df["D_error"], linewidth = 1, color = "#6baed6")
ax1.plot(msd_d_df["t"]/1000, msd_d_df["Mean_D"] + msd_d_df["D_error"], linewidth = 1, color = "#6baed6")
ax1.fill_between(msd_d_df["t"]/1000, msd_d_df["Mean_D"] + msd_d_df["D_error"], msd_d_df["Mean_D"] - msd_d_df["D_error"], color = "#9ecae1", alpha = 0.2)
ax.annotate("", xy=(0.7, 0.68), xycoords = "axes fraction",
xytext=(0.85, 0.68), textcoords='axes fraction',
arrowprops=dict(arrowstyle="->", color = "#525252"))
ax.annotate("", xy=(0.23, 0.40), xycoords = "axes fraction",
xytext=(0.08, 0.40), textcoords='axes fraction',
arrowprops=dict(arrowstyle="->", color = color))
xmin, xmax = ax.get_xlim()
ymin, ymax = ax.get_ylim()
ax.text(xmin + 0.35*(xmax-xmin), ymin + 0.92*(ymax - ymin), "%.2e cm$^2$ s$^{-1}$"%(linear_model[0]/60000), ha = "center", va = "center")
ax.set_ylabel('MSD (Å$^2$)')
ax.set_xlabel('$t$ (ps)')
ax1.set_ylabel('$D$ (cm$^2$ s$^{-1}$)', color = color)
ax1.tick_params(axis='y', color = color, labelcolor = color)
ax1.spines['right'].set_color(color)
ax1.ticklabel_format(axis='y', style='sci', scilimits=[-4,4], useMathText=True)
plt.savefig("test/cage1-ave_msd-d-500K.jpg", dpi =600)
plt.show()